import numpy as np
from epymorph.kit import *
from epymorph.adrio import acs5
from epymorph.initializer import ProportionalEstimating space- and time-varying transmission rate from data
Scenario
It is common that we want to account for and quanitfy heterogeneity in transmission rates among localities to better understand the regional picture of pathogen spread. Therefore, we may have a spatial model with multiple nodes, and we want to understand from data how the transmission rate varies among localities. We may also want to quantify this spatial heterogeneity while also accounting for host movement among localities. In epymorph, we have built convenient parameter estimation functionality that allows us to estimate location- and time-varying parameters. In this scenario, we will explore how to set up the model and particle filter simulation, and we will demonstrate the critical role of the particle filter resampling protocol for optimizing the power of our analysis.
Exercise 1
We will first demonstrate why the single-node particle filter from the ealier vignette is insufficient for higher-dimentional problems. First, we will develop a multi-node simulation that does not include the effects of movement.
states = ["AZ", "CO", "NM", "UT"]
rume = SingleStrataRUME.build(
ipm=ipm.SIRH(),
mm=mm.No(),
scope=StateScope.in_states(states, year=2015),
init=Proportional(
ratios=np.broadcast_to(
np.array([9999, 1, 0, 0], dtype=np.int64), shape=(len(states), 4)
)
),
time_frame=TimeFrame.of("2022-09-15", 7 * 26 + 1),
params={
"beta": 0.3, # Placeholder value
"gamma": 0.2,
"xi": 1 / 365,
"hospitalization_prob": 200 / 100_000,
"hospitalization_duration": 5.0,
"population": acs5.Population(),
},
)from epymorph.parameter_fitting.distribution import Uniform
from epymorph.parameter_fitting.utils.parameter_estimation import EstimateParameters
from epymorph.parameter_fitting.dynamics import GeometricBrownianMotion
params_space = {
"beta": EstimateParameters.TimeVarying(
distribution=Uniform(a=0.1, b=0.8),
dynamics=GeometricBrownianMotion(volatility=0.1),
),
}from epymorph.adrio.cdc import InfluenzaStateHospitalization
from epymorph.parameter_fitting.likelihood import Poisson
from epymorph.parameter_fitting.utils.observations import ModelLink, Observations
from epymorph.time import EveryNDays
observations = Observations(
source=InfluenzaStateHospitalization(),
model_link=ModelLink(
quantity=rume.ipm.select.events("I->H"),
time=rume.time_frame.select.all().group(EveryNDays(7)).agg(),
geo=rume.scope.select.all(),
),
likelihood=Poisson(),
)from epymorph.parameter_fitting.filter.particle_filter import ParticleFilter
filter_type = ParticleFilter(num_particles=300)from epymorph.parameter_fitting.particlefilter_simulation import FilterSimulation
sim = FilterSimulation(
rume=rume,
observations=observations,
filter_type=filter_type,
params_space=params_space,
)import matplotlib.pyplot as plt
time_axis = range(len(sim.cases))
n_nodes = sim.cases.shape[1]
for i_node in range(n_nodes):
plt.bar(
time_axis,
sim.cases[:, i_node],
bottom=sim.cases[:, 0:i_node].sum(axis=1),
label=rume.scope.labels[i_node],
)
plt.title("Stacked bar chart of weekly hospitalizations")
plt.grid(True)
plt.xlabel("Time (weeks)")
plt.ylabel("Hospitalizations")
plt.legend()
plt.show()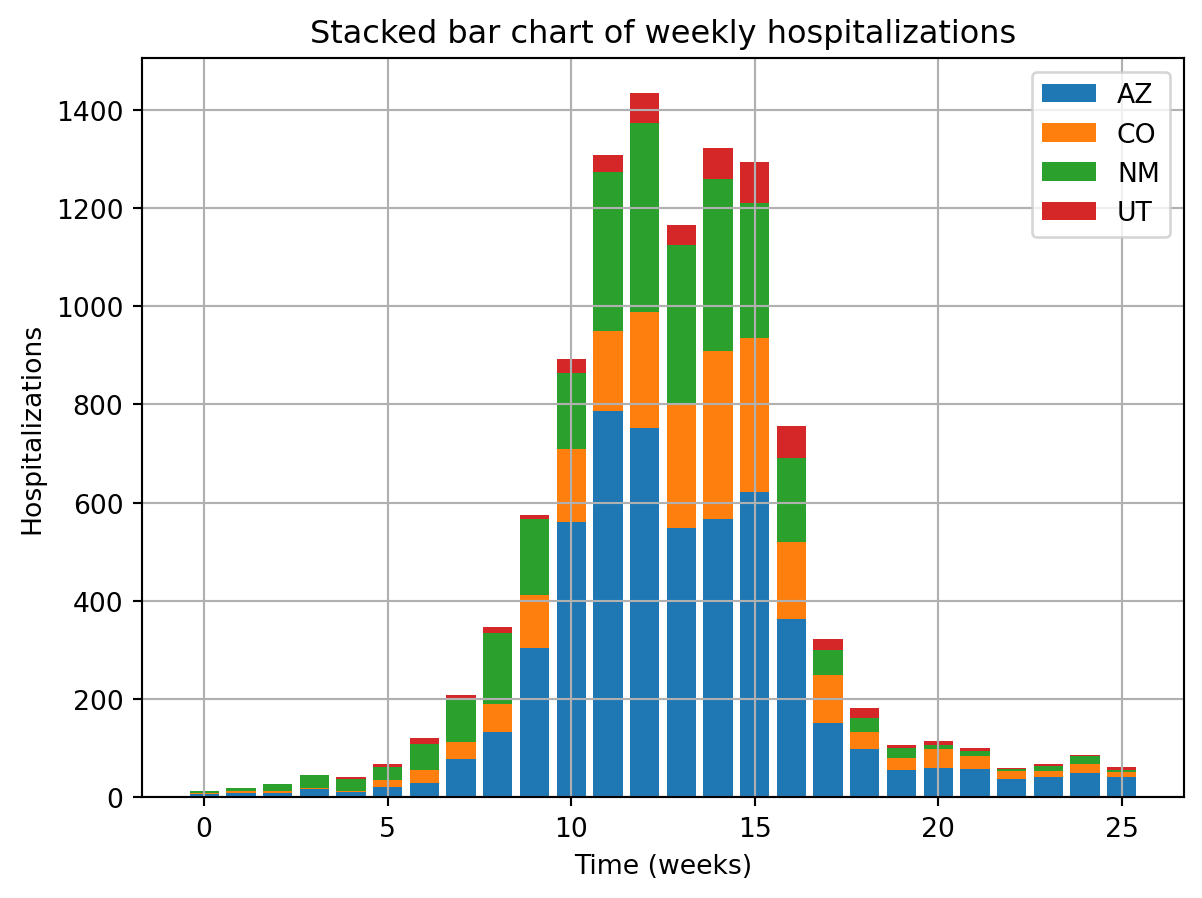
fig, axs = plt.subplots(2, 2, sharey=True)
axs = np.ravel(axs)
for i_ax in range(len(axs)):
ax = axs[i_ax]
ax.bar(
time_axis,
sim.cases[:, i_ax],
)
ax.set_title(f"Weekly Hosp. in {rume.scope.labels[i_ax]}")
ax.set_xlabel("Time (weeks)")
ax.set_ylabel("Hospitalizations")
plt.tight_layout()
plt.show()
output = sim.run()fig, axs = plt.subplots(2, 2, sharey=True)
key = "beta"
key_quantiles = np.array(output.param_quantiles[key])
t = np.arange(0, len(key_quantiles))
axs = np.ravel(axs)
for i_ax in range(len(axs)):
ax = axs[i_ax]
ax.fill_between(
t,
key_quantiles[:, 3, i_ax],
key_quantiles[:, 22 - 3, i_ax],
facecolor="blue",
alpha=0.2,
label="Quantile Range (10th to 90th)",
)
ax.fill_between(
t,
key_quantiles[:, 6, i_ax],
key_quantiles[:, 22 - 6, i_ax],
facecolor="blue",
alpha=0.4,
label="Quantile Range (25th to 75th)",
)
ax.plot(
t,
key_quantiles[:, 11, i_ax],
color="red",
label="Median (50th Percentile)",
)
ax.set_title(f"Estimate of '{key}' in {rume.scope.labels[i_ax]}")
ax.set_xlabel("Time (weeks)")
ax.set_ylabel("Quantiles")
ax.grid(True)
plt.legend()
plt.tight_layout()
plt.show()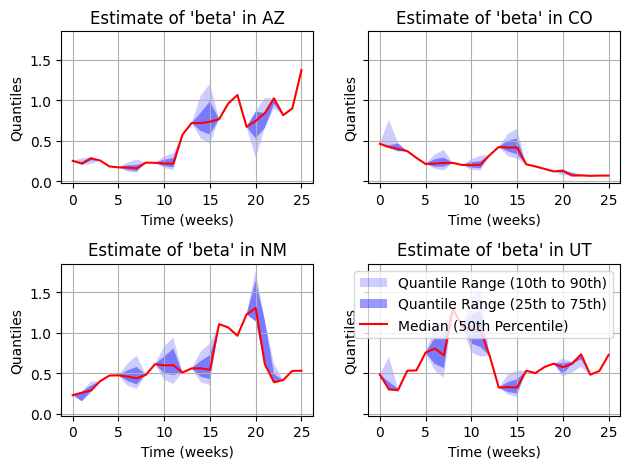
YIKES! What we see is catastrophic collapse of the particle filter. The dimentionality is too big for this type of resampler. But, we can fix this issue.
Exercise 2
The particle filter can suffer from the “curse of dimensionality”, meaning that as the number of nodes increases, the number of particles needed can increase to a point where a normal particle filter is impractical. In this case there we developed an alternative resampler called ResamplingByNode which makes a small approximation (localization technique) to greatly improve performance in multi-node systems.
from epymorph.parameter_fitting.utils.resampler import ResamplingByNode
filter_type = ParticleFilter(num_particles=300, resampler=ResamplingByNode)sim = FilterSimulation(
rume=rume,
observations=observations,
filter_type=filter_type,
params_space=params_space,
)output = sim.run()fig, axs = plt.subplots(2, 2, sharey=True)
key = "beta"
key_quantiles = np.array(output.param_quantiles[key])
t = np.arange(0, len(key_quantiles))
axs = np.ravel(axs)
for i_ax in range(len(axs)):
ax = axs[i_ax]
ax.fill_between(
t,
key_quantiles[:, 3, i_ax],
key_quantiles[:, 22 - 3, i_ax],
facecolor="blue",
alpha=0.2,
label="Quantile Range (10th to 90th)",
)
ax.fill_between(
t,
key_quantiles[:, 6, i_ax],
key_quantiles[:, 22 - 6, i_ax],
facecolor="blue",
alpha=0.4,
label="Quantile Range (25th to 75th)",
)
ax.plot(
t,
key_quantiles[:, 11, i_ax],
color="red",
label="Median (50th Percentile)",
)
ax.set_title(f"Estimate of '{key}' in {rume.scope.labels[i_ax]}")
ax.set_xlabel("Time (weeks)")
ax.set_ylabel("Quantiles")
ax.grid(True)
plt.legend()
plt.tight_layout()
plt.show()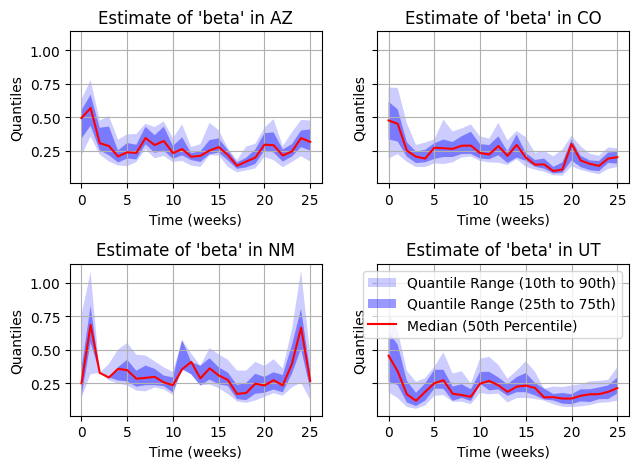
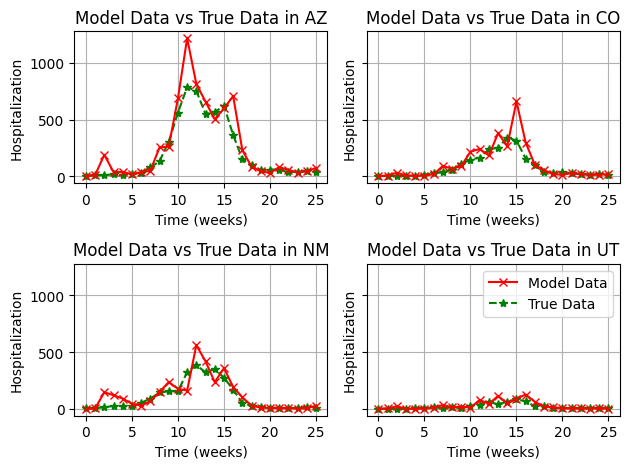
This is much better. We can still very minor issues with particle filter collapse in some areas, but in general, the estimation is much more robust. It would be recommended to increase the number of particles to make sure we don’t see any collapse.
fig, axs = plt.subplots(2, 2, sharey=True)
axs = np.ravel(axs)
for i_ax in range(len(axs)):
ax = axs[i_ax]
ax.plot(
output.model_data[:, i_ax],
label="Model Data",
color="red",
linestyle="-",
zorder=10,
marker="x",
)
ax.plot(
output.true_data[:, i_ax],
label="True Data",
color="green",
linestyle="--",
zorder=9,
marker="*",
)
ax.set_xlabel("Time (weeks)")
ax.set_ylabel("Hospitalization")
ax.set_title(f"Model Data vs True Data in {rume.scope.labels[i_ax]}")
ax.grid(True)
plt.legend()
plt.tight_layout()
plt.show()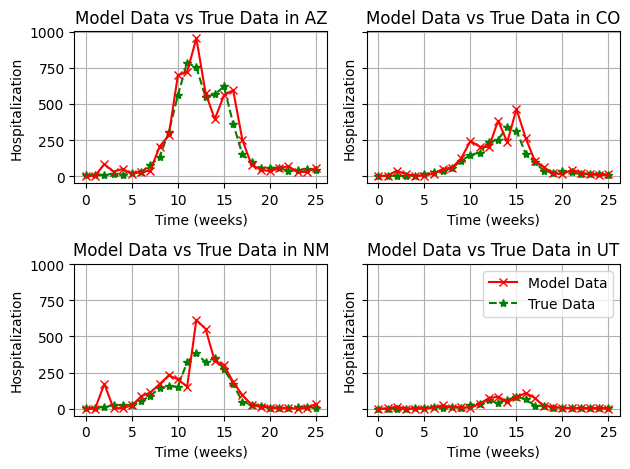
Exercise 3
In this final example, we will fit the State-specific transmission rates, but our model will also include the effects of inter-state commuter movement.
from epymorph.adrio import commuting_flows
rume = SingleStrataRUME.build(
ipm=ipm.SIRH(),
mm=mm.Pei(),
scope=StateScope.in_states(states, year=2015),
init=Proportional(
ratios=np.broadcast_to(
np.array([9999, 1, 0, 0], dtype=np.int64), shape=(len(states), 4)
)
),
time_frame=TimeFrame.of("2022-09-15", 7 * 26 + 1),
params={
"beta": 0.3, # Placeholder value
"gamma": 0.2,
"xi": 1 / 365,
"hospitalization_prob": 200 / 100_000,
"hospitalization_duration": 5.0,
"population": acs5.Population(),
"commuters": commuting_flows.Commuters(),
},
)Note: The commuters ADRIO takes some time to evaluate, even when the data is cached. This would be a problem if we passed this RUME directly into the particle filter, which runs hundreds of simulations. To bypass this, we’re going to precompute the commuter matrix, which is a pretty advanced technique, but saves a lot of compute time. A user could run the particle filter without this step, but it would take a lot longer to run.
import dataclasses
from epymorph.attribute import AbsoluteName, NamePattern
name = AbsoluteName(strata="gpm:all", module="mm", id="commuters")
attr_def = rume.requirements[name]
# Precompute the commuter matrix.
rng = np.random.default_rng(seed=1)
data_resolver = rume.evaluate_params(rng=rng)
commuter_matrix = data_resolver.resolve(name, attr_def)
# Create a copy of the RUME, but with the commuter matrix replaced.
params_copy = dict(rume.params)
params_copy[NamePattern.of(name.id)] = commuter_matrix
rume_copy = dataclasses.replace(rume, params=params_copy)observations = Observations(
source=InfluenzaStateHospitalization(),
model_link=ModelLink(
quantity=rume.ipm.select.events("I->H"),
time=rume.time_frame.select.all().group(EveryNDays(7)).agg(),
geo=rume.scope.select.all(),
),
likelihood=Poisson(),
)sim = FilterSimulation(
rume=rume_copy,
observations=observations,
filter_type=filter_type,
params_space=params_space,
)output = sim.run(rng=rng)fig, axs = plt.subplots(2, 2, sharey=True)
key = "beta"
key_quantiles = np.array(output.param_quantiles[key])
t = np.arange(0, len(key_quantiles))
axs = np.ravel(axs)
for i_ax in range(len(axs)):
ax = axs[i_ax]
ax.fill_between(
t,
key_quantiles[:, 3, i_ax],
key_quantiles[:, 22 - 3, i_ax],
facecolor="blue",
alpha=0.2,
label="Quantile Range (10th to 90th)",
)
ax.fill_between(
t,
key_quantiles[:, 6, i_ax],
key_quantiles[:, 22 - 6, i_ax],
facecolor="blue",
alpha=0.4,
label="Quantile Range (25th to 75th)",
)
ax.plot(
t,
key_quantiles[:, 11, i_ax],
color="red",
label="Median (50th Percentile)",
)
ax.set_title(f"Estimate of '{key}' in {rume.scope.labels[i_ax]}")
ax.set_xlabel("Time (weeks)")
ax.set_ylabel("Quantiles")
ax.grid(True)
plt.legend()
plt.tight_layout()
plt.show()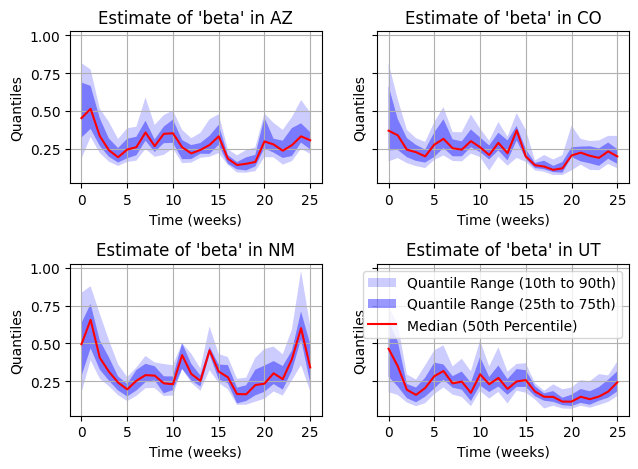
We can see that the estimates of transmission rate are not very different, such that movement at this spatial scale (State-level) likely does not have a huge effect on the local and regional dynamics.
fig, axs = plt.subplots(2, 2, sharey=True)
axs = np.ravel(axs)
for i_ax in range(len(axs)):
ax = axs[i_ax]
ax.plot(
output.model_data[:, i_ax],
label="Model Data",
color="red",
linestyle="-",
zorder=10,
marker="x",
)
ax.plot(
output.true_data[:, i_ax],
label="True Data",
color="green",
linestyle="--",
zorder=9,
marker="*",
)
ax.set_xlabel("Time (weeks)")
ax.set_ylabel("Hospitalization")
ax.set_title(f"Model Data vs True Data in {rume.scope.labels[i_ax]}")
ax.grid(True)
plt.legend()
plt.tight_layout()
plt.show()