import matplotlib.pyplot as plt
import numpy as np
from epymorph.kit import *
from epymorph.adrio import acs5Multi-node Simulations with Movement Models
SCENARIO:
Imagine a new respiratory pathogen emerges in Maricopa County, AZ where 5 people are initially infected. In an effort to control the outbreak, local public health officials across the state have asked us for guidance on how the pathogen might spread to different counties. We therefore need to consider human movement patterns in our model to understand spatial spread. We will build a County-level spatial model with epymorph, such that each node of our model represents the population in a single county. The model then assumes that the populations within the county interact homogeneously. Each node is interconnected through human movement, which affects the regional spread of the pathogen.
The exercises in this vignette show you how to add nodes and movement between nodes to your disease models in epymorph and will explore how varying how we model movement (i.e., the assumptions that govern movement flows) might impact disease dynamics.
Exercise 1
my_scope = CountyScope.in_states(['AZ'], year=2022)
my_scope.labels
# You can also see their FIPS code:
# my_scope.node_idsarray(['Apache, AZ', 'Cochise, AZ', 'Coconino, AZ', 'Gila, AZ',
'Graham, AZ', 'Greenlee, AZ', 'La Paz, AZ', 'Maricopa, AZ',
'Mohave, AZ', 'Navajo, AZ', 'Pima, AZ', 'Pinal, AZ',
'Santa Cruz, AZ', 'Yavapai, AZ', 'Yuma, AZ'], dtype='<U14')from epymorph.data.ipm.sirs import SIRS
sirs_ipm = SIRS()sirs_ipm.diagram()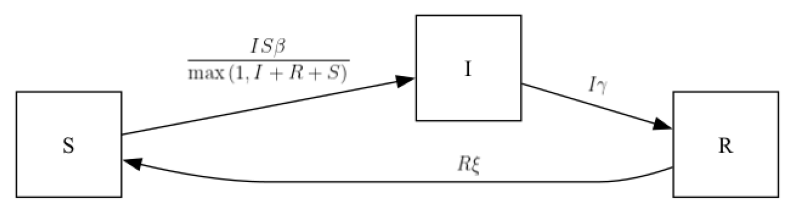
from epymorph.data.mm.flat import Flat
flat_mm = Flat()# Maricopa is the 8th node in the scope (i.e., index 7)
my_init = init.SingleLocation(location=7, seed_size=5)time = TimeFrame.of("2022-01-01", duration_days=190)comm_prop = 0.15beta = 0.4
gam = 1 / 4
xi = 1 / 200rume = SingleStrataRUME.build(
scope=my_scope,
ipm=sirs_ipm,
mm=flat_mm,
init=my_init,
time_frame=time,
params={
'beta': beta,
'gamma': gam,
'xi': xi,
'population': acs5.Population(),
'commuter_proportion': comm_prop,
}
)
# Print a description of params
print(rume.params_description())ipm::beta (type: float, shape: TxN)
infectivity
ipm::gamma (type: float, shape: TxN)
progression from infected to recovered
ipm::xi (type: float, shape: TxN)
progression from recovered to susceptible
mm::population (type: int, shape: N)
The total population at each node.
mm::commuter_proportion (type: float, shape: S, default: 0.1)
The proportion of the total population which commutes.
init::population (type: int, shape: N)
The population at each geo node.
- Run your simulation
sim = BasicSimulator(rume)
with sim_messaging(live = False):
out = sim.run(
rng_factory=default_rng(5),
)Loading gpm:all::mm::population (epymorph.adrio.acs5.Population):
|####################| 100% (0.736s)
Running simulation (BasicSimulator):
• 2022-01-01 to 2022-07-09 (190 days)
• 15 geo nodes
|####################| 100%
Runtime: 0.918sout.plot.line(
geo=out.rume.scope.select.all(),
time=out.rume.time_frame.select.all().group("day").agg(),
quantity=out.rume.ipm.select.compartments("I"),
title="Infectious in AZ Counties",
legend="outside",
)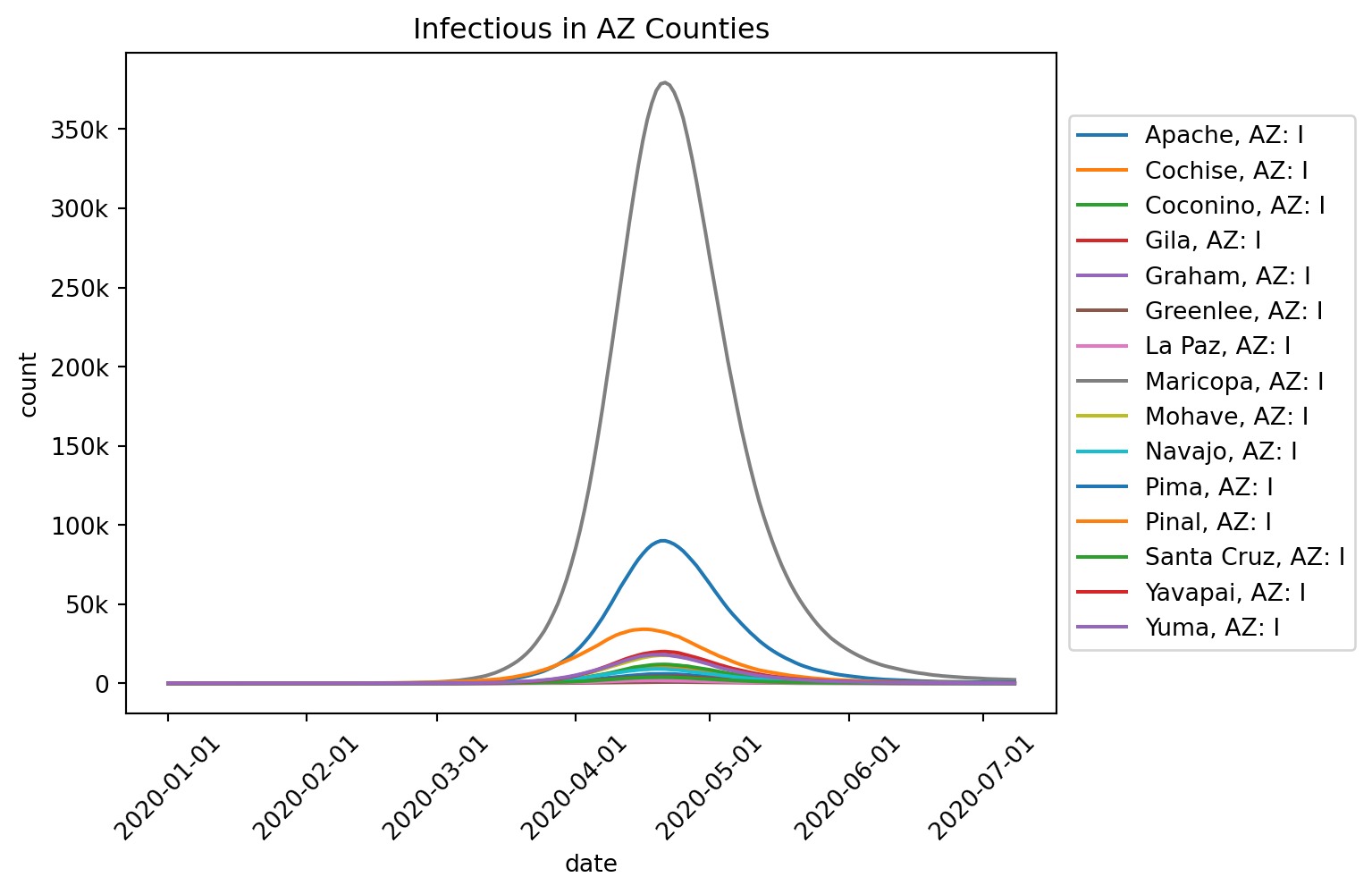
out.table.quantiles(
quantiles=[1.0],
geo=out.rume.scope.select.all(),
time=out.rume.time_frame.select.all().group("day").agg(),
quantity=out.rume.ipm.select.compartments("I"),
)| geo | quantity | 1.0 | |
|---|---|---|---|
| 0 | Apache, AZ | I | 5611.0 |
| 1 | Cochise, AZ | I | 10883.0 |
| 2 | Coconino, AZ | I | 12227.0 |
| 3 | Gila, AZ | I | 4653.0 |
| 4 | Graham, AZ | I | 3218.0 |
| 5 | Greenlee, AZ | I | 827.0 |
| 6 | La Paz, AZ | I | 1404.0 |
| 7 | Maricopa, AZ | I | 372942.0 |
| 8 | Mohave, AZ | I | 18902.0 |
| 9 | Navajo, AZ | I | 9038.0 |
| 10 | Pima, AZ | I | 91661.0 |
| 11 | Pinal, AZ | I | 38199.0 |
| 12 | Santa Cruz, AZ | I | 4177.0 |
| 13 | Yavapai, AZ | I | 20833.0 |
| 14 | Yuma, AZ | I | 18275.0 |
def log10_transform(data_df):
# we have to be careful to avoid log(0)!
log_value = data_df["data"].apply(lambda x: np.log10(x) if x > 0 else nan)
return data_df.assign(data=log_value)
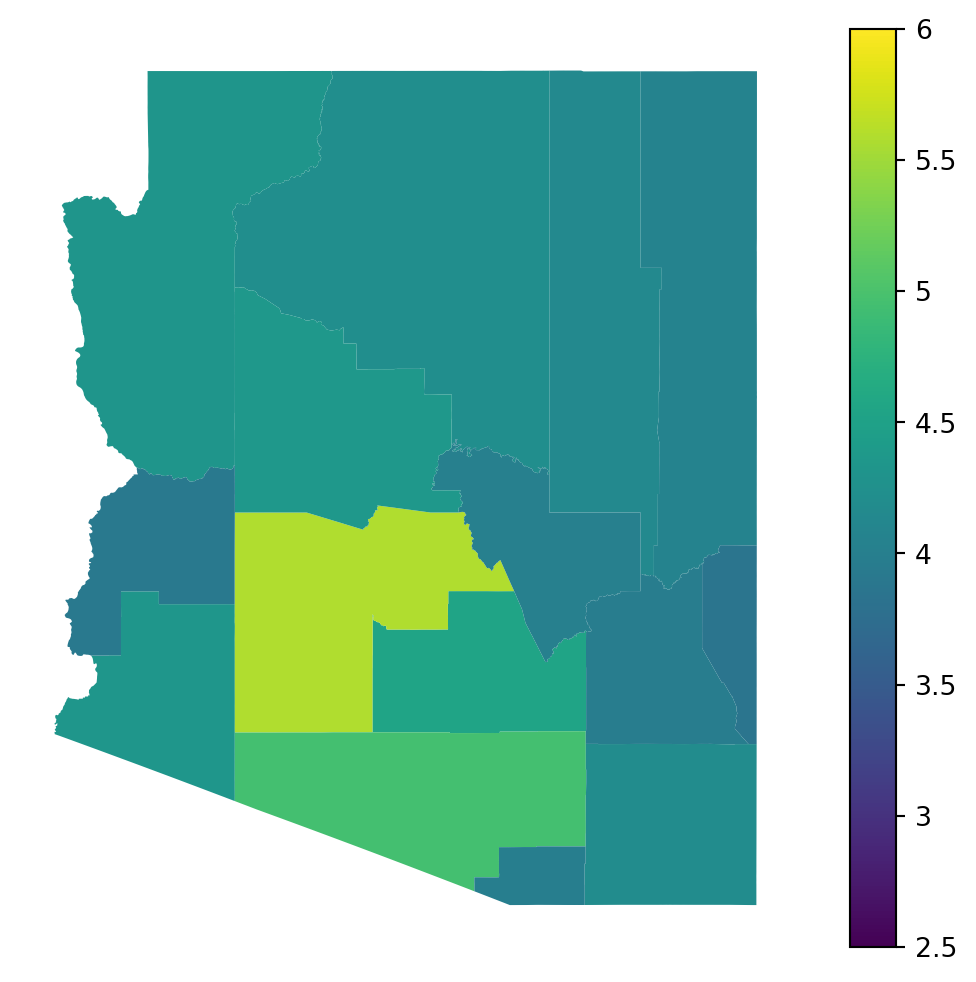
out.map.choropleth(
geo=rume.scope.select.all(),
time=rume.time_frame.select.rangex("2022-04-01", "2022-05-01").agg(compartments="max"),
quantity=rume.ipm.select.compartments("I"),
transform=log10_transform,
vmax = 6.0,
vmin = 2.5,
)
Exercise 2
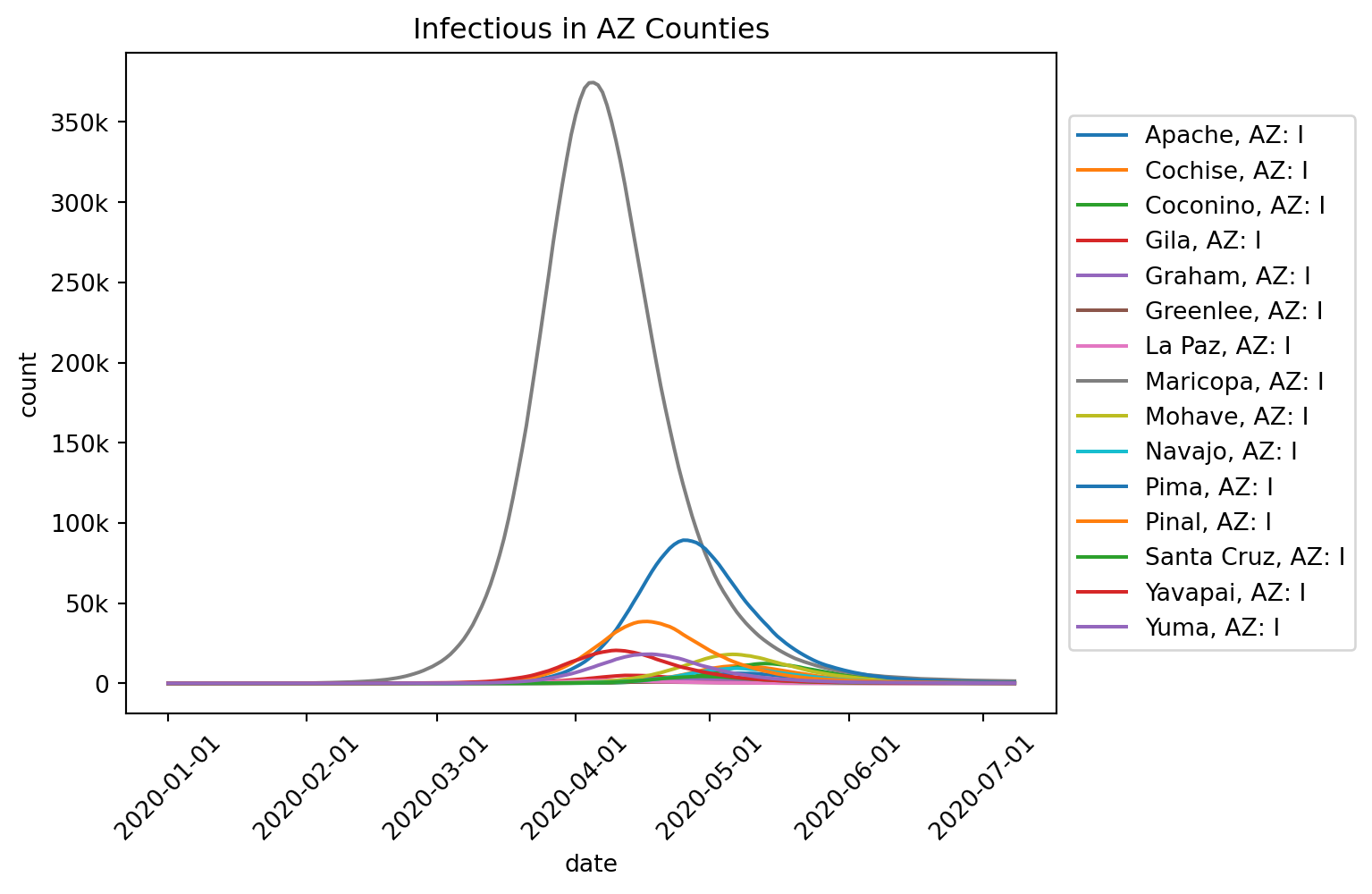
In Exercise 1, we used a “flat” movement model, in which individuals could move equally randomly to any other node. This led to more or less syncronous outbreaks across space, which is of course not very realistic. In this Exercise, we’ll use more realistic movement models.
Note that this new MM requires a calculation of centroids, but this can be done using the us_tiger ADRIO. Additionally, we need a parameter ‘phi’ which modulates the effect of distance in the probability kernel. On your own, you can test how ‘phi’ influences the dynamics.
from epymorph.data.mm.centroids import Centroids
from epymorph.adrio import us_tiger
rume = SingleStrataRUME.build(
scope=my_scope,
ipm=sirs_ipm,
mm=Centroids(),
init=my_init,
time_frame=time,
params={
#For IPM
'beta': beta,
'gamma': gam,
'xi': xi,
#For MM
'population': acs5.Population(),
'commuter_proportion': comm_prop,
'centroid': us_tiger.InternalPoint(),
'phi': 20.0,
}
)
print(rume.params_description())ipm::beta (type: float, shape: TxN)
infectivity
ipm::gamma (type: float, shape: TxN)
progression from infected to recovered
ipm::xi (type: float, shape: TxN)
progression from recovered to susceptible
mm::population (type: int, shape: N)
The total population at each node.
mm::centroid (type: [(longitude, float), (latitude, float)], shape: N)
The centroids for each node as (longitude, latitude) tuples.
mm::phi (type: float, shape: S, default: 40.0)
Influences the distance that movers tend to travel.
mm::commuter_proportion (type: float, shape: S, default: 0.1)
The proportion of the total population which commutes.
init::population (type: int, shape: N)
The population at each geo node.
sim = BasicSimulator(rume)
with sim_messaging(live = False):
out = sim.run(
rng_factory=default_rng(5),
)Loading gpm:all::mm::population (epymorph.adrio.acs5.Population):
|####################| 100% (1.241s)
Running simulation (BasicSimulator):
• 2022-01-01 to 2022-07-09 (190 days)
• 15 geo nodes
|####################| 100%
Runtime: 0.820sout.plot.line(
geo=out.rume.scope.select.all(),
time=out.rume.time_frame.select.all().group("day").agg(),
quantity=out.rume.ipm.select.compartments("I"),
title="Infectious in AZ Counties",
legend="outside",
)
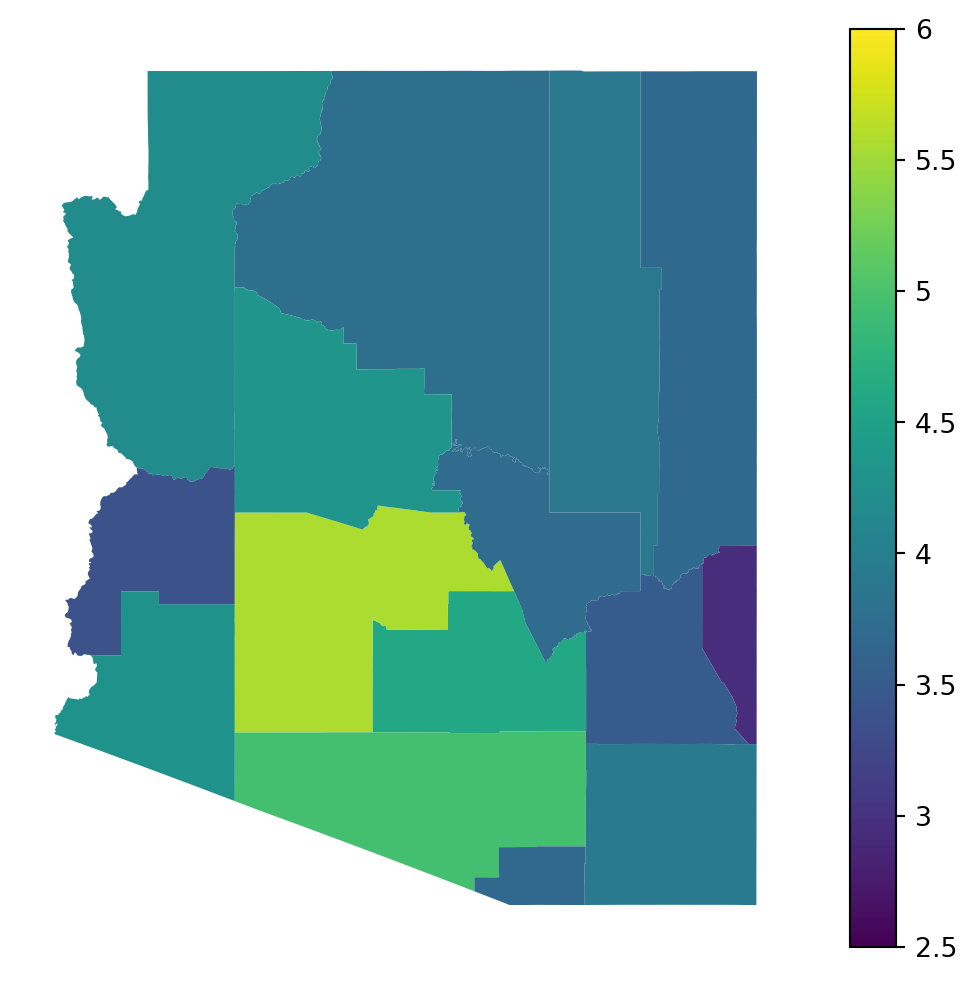
out.map.choropleth(
geo=rume.scope.select.all(),
time=rume.time_frame.select.rangex("2022-04-01", "2022-05-01").agg(compartments="max"),
quantity=rume.ipm.select.compartments("I"),
transform=log10_transform,
vmax = 6.0,
vmin = 2.5,
)
Exercise 3
Finally, use a movement model that is based on real data on human behavior. Specifically, we will use a commuter-style movement model created from a model published by Pei et al. This model requires a commuter matrix, and we will use mobility data from the ACS5 commuting flows, which are at the County level. Notice that for the MM, we will use some control parameters with their defaults.
from epymorph.data.mm.pei import Pei as CommuterMM
from epymorph.adrio import commuting_flows
rume = SingleStrataRUME.build(
scope=my_scope,
ipm=sirs_ipm,
mm=CommuterMM(),
init=my_init,
time_frame=time,
params={
#For IPM
'beta': beta,
'gamma': gam,
'xi': xi,
#For MM
'commuters': commuting_flows.Commuters(),
# General:
'population': acs5.Population(),
}
)
print(rume.params_description())ipm::beta (type: float, shape: TxN)
infectivity
ipm::gamma (type: float, shape: TxN)
progression from infected to recovered
ipm::xi (type: float, shape: TxN)
progression from recovered to susceptible
mm::commuters (type: int, shape: NxN)
A node-to-node commuters marix.
mm::move_control (type: float, shape: S, default: 0.9)
A factor which modulates the number of commuters by conducting a
binomial draw with this probability and the expected commuters
from the commuters matrix.
mm::theta (type: float, shape: S, default: 0.1)
A factor which allows for randomized movement by conducting a
poisson draw with this factor times the average number of
commuters between two nodes from the commuters matrix.
init::population (type: int, shape: N)
The population at each geo node.
sim = BasicSimulator(rume)
with sim_messaging(live = False):
out = sim.run(
rng_factory=default_rng(5),
)Loading gpm:all::mm::commuters (epymorph.adrio.commuting_flows.Commuters):
|####################| 100% (10.920s)
Loading gpm:all::init::population (epymorph.adrio.acs5.Population):
|####################| 100% (0.876s)
Running simulation (BasicSimulator):
• 2022-01-01 to 2022-07-09 (190 days)
• 15 geo nodes
|####################| 100%
Runtime: 0.997sout.plot.line(
geo=out.rume.scope.select.all(),
time=out.rume.time_frame.select.all().group("day").agg(),
quantity=out.rume.ipm.select.compartments("I"),
title="Infectious in AZ Counties",
legend="outside",
)