from epymorph.adrio import acs5, csv
from epymorph.data.ipm.sirh import SIRH
from epymorph.data.mm.no import No
from epymorph.initializer import Proportional
import numpy as np
from epymorph.kit import *Estimating time-varying transmission rate from data
SCENARIO:
A particle filter is an algorithm which can use data to estimate a parameter, such as the transmission rate. The particle filter accomplishes this by combining the data with a mathematical model.
This vignette will demonstrate how to use epymorph’s built-in particle filter to estimate a time-varying transmission rate (beta) using weekly influenza hospitalization data. In the first exercise, we will define our model setup and generate synthetic data (fake data used for testing) which will be used to validate the particle filter. In the second exercise, we will setup the particle filter and run it on the synthetic data we generated in the first exercise. By validating the particle filter setup on synthetic data first, we can diagnose any issues before moving on to real data. In the third exercise, we will run the particle filter on real influenza hospitalization data. Because we will have already tested the particle filter on synthetic data, this step will be as easy as swapping out the synthetic data for the real data.
Exercise 1
In this first exercise, we will setup out mathematical model and generate synthetic data.
num_weeks = 26
duration = 7 * num_weeks + 1
t = np.arange(0, duration)
true_beta = 0.05 * np.cos(t * 2 * np.pi / (365)) + 0.25import matplotlib.pyplot as plt
plt.plot(t, true_beta)
plt.ylabel("beta")
plt.xlabel("time (days)")
plt.title("The transmission rate (beta) over time")
plt.show()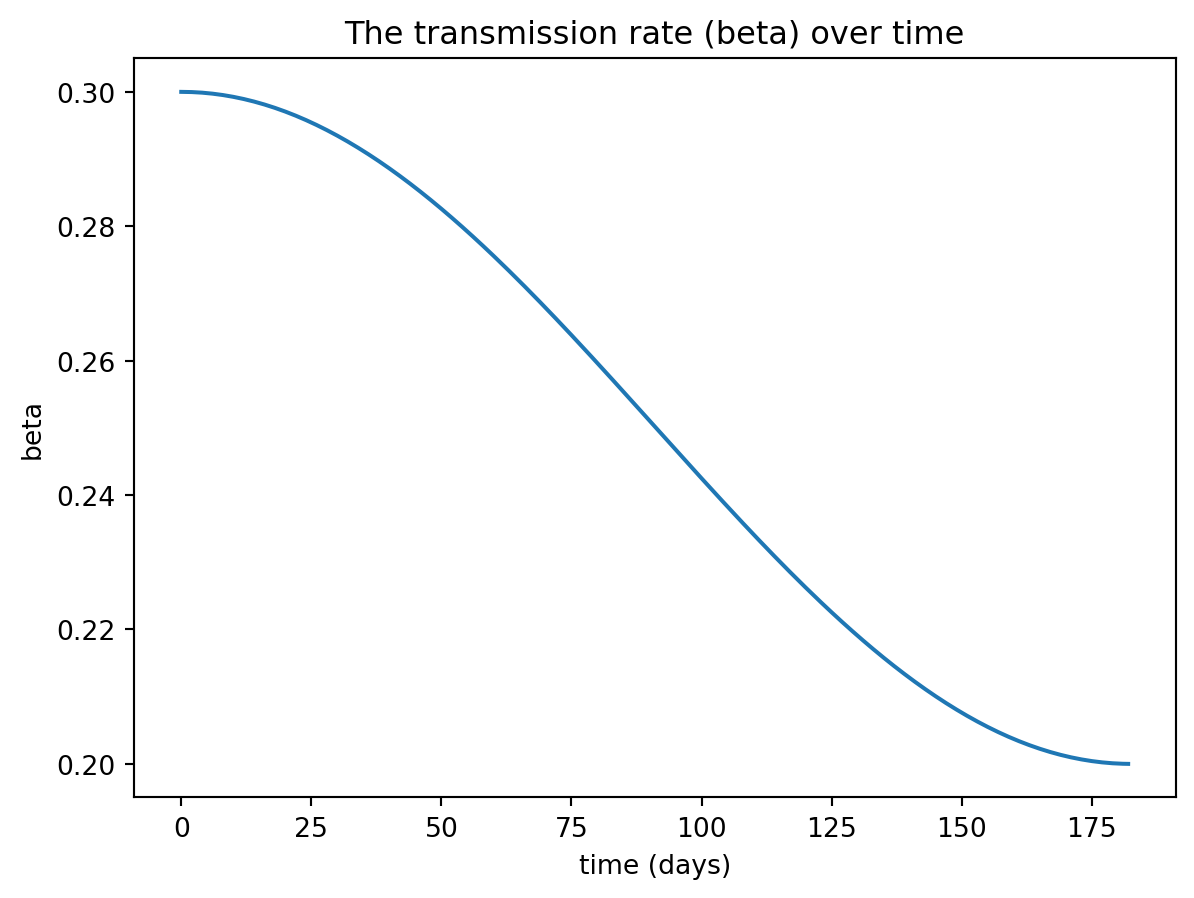
my_ipm = SIRH()
my_ipm.diagram()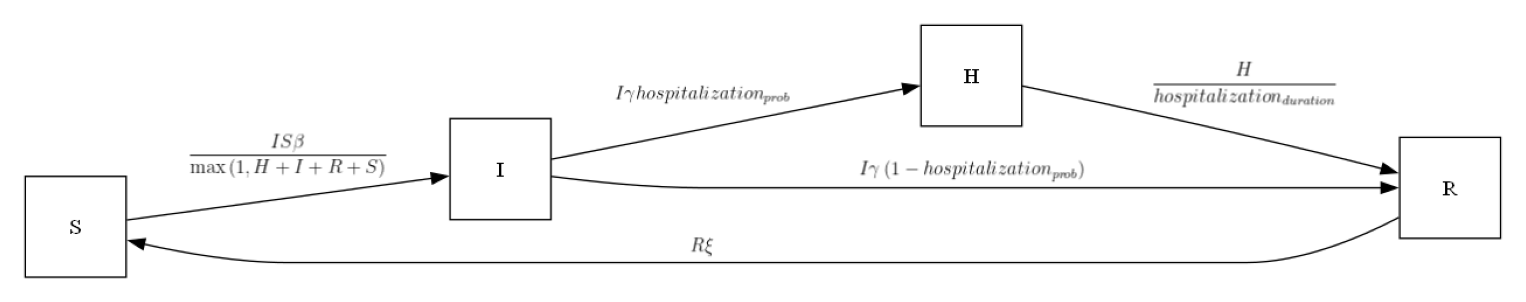
rume = SingleStrataRUME.build(
ipm=SIRH(),
mm=No(),
scope=StateScope.in_states(["AZ"], year=2015),
init=Proportional(ratios=np.array([9999, 1, 0, 0], dtype=np.int64)),
time_frame=TimeFrame.of("2022-09-15", 7 * 26 + 1),
params={
"beta": true_beta,
"gamma": 0.2,
"xi": 1 / 365,
"hospitalization_prob": 200 / 100_000,
"hospitalization_duration": 5.0,
"population": acs5.Population(),
},
)rng = np.random.default_rng(seed=1)
sim = BasicSimulator(rume)
with sim_messaging(live = False):
out = sim.run(rng_factory=(lambda: rng))Loading gpm:all::init::population (epymorph.adrio.acs5.Population):
|####################| 100% (0.688s)
Running simulation (BasicSimulator):
• 2022-09-15 to 2023-03-16 (183 days)
• 1 geo nodes
|####################| 100%
Runtime: 0.052simport tempfile
from epymorph.time import EveryNDays
from epymorph.tools.data import munge
quantity_selection = rume.ipm.select.events("I->H")
time_selection = rume.time_frame.select.all().group(EveryNDays(7)).agg()
geo_selection = rume.scope.select.all()
cases_df = munge(
out,
quantity=quantity_selection,
time=time_selection,
geo=geo_selection,
)
# We'll use a temporary file here,
# but typically you would use a normal file.
temp_file = tempfile.NamedTemporaryFile(suffix=".csv", delete=False).name
cases_df.to_csv(temp_file, index=False)Exercise 2
In this second exercise, we will run the particle filter on the synthetic data we generated and compare the estimate of beta produced by the particle filter to the true values of beta which were used to generate the data.
from epymorph.time import DateRange
csvadrio = csv.CSVFileTxN(
file_path=temp_file,
dtype=int,
key_col=1,
key_type="geoid",
time_col=0,
data_col=2,
skiprows=1,
date_range=rume.time_frame.to_date_range(),
)from epymorph.parameter_fitting.utils.observations import ModelLink
model_link=ModelLink(
quantity=quantity_selection,
time=time_selection,
geo=geo_selection,
)from epymorph.parameter_fitting.likelihood import Poisson
from epymorph.parameter_fitting.utils.observations import Observations
observations = Observations(
source=csvadrio,
model_link=model_link,
likelihood=Poisson(),
)from epymorph.parameter_fitting.filter.particle_filter import ParticleFilter
filter_type = ParticleFilter(num_particles=200)from epymorph.parameter_fitting.distribution import Uniform
from epymorph.parameter_fitting.dynamics import GeometricBrownianMotion
from epymorph.parameter_fitting.utils.parameter_estimation import EstimateParameters
params_space = {
"beta": EstimateParameters.TimeVarying(
distribution=Uniform(a=0.05, b=0.5),
dynamics=GeometricBrownianMotion(volatility=0.05),
),
}from epymorph.parameter_fitting.particlefilter_simulation import FilterSimulation
sim = FilterSimulation(
rume=rume,
observations=observations,
filter_type=filter_type,
params_space=params_space,
)plt.bar(range(len(sim.cases)), sim.cases[:, 0])
plt.title("Observations")
plt.xlabel("Time (weeks)")
plt.ylabel("Hospitalizations")
plt.grid(True)
plt.show()
output = sim.run(rng=rng)key = "beta"
key_quantiles = np.array(output.param_quantiles[key])
plt.fill_between(
np.arange(0, len(key_quantiles)),
key_quantiles[:, 3, 0],
key_quantiles[:, 22 - 3, 0],
facecolor="blue",
alpha=0.2,
label="Quantile Range (10th to 90th)",
)
plt.fill_between(
np.arange(0, len(key_quantiles)),
key_quantiles[:, 6, 0],
key_quantiles[:, 22 - 6, 0],
facecolor="blue",
alpha=0.4,
label="Quantile Range (25th to 75th)",
)
plt.plot(
np.arange(0, len(key_quantiles)),
key_quantiles[:, 11, 0],
color="red",
label="Median (50th Percentile)",
)
obs = np.arange(0, len(key_quantiles))
plt.plot(
1/7*t,
true_beta,
"k--",
label="Truth"
)
plt.title(f"Estimate of '{key}'")
plt.xlabel("Time (weeks)")
plt.ylabel("Quantiles")
plt.legend(loc="upper right")
plt.grid(True)
plt.show()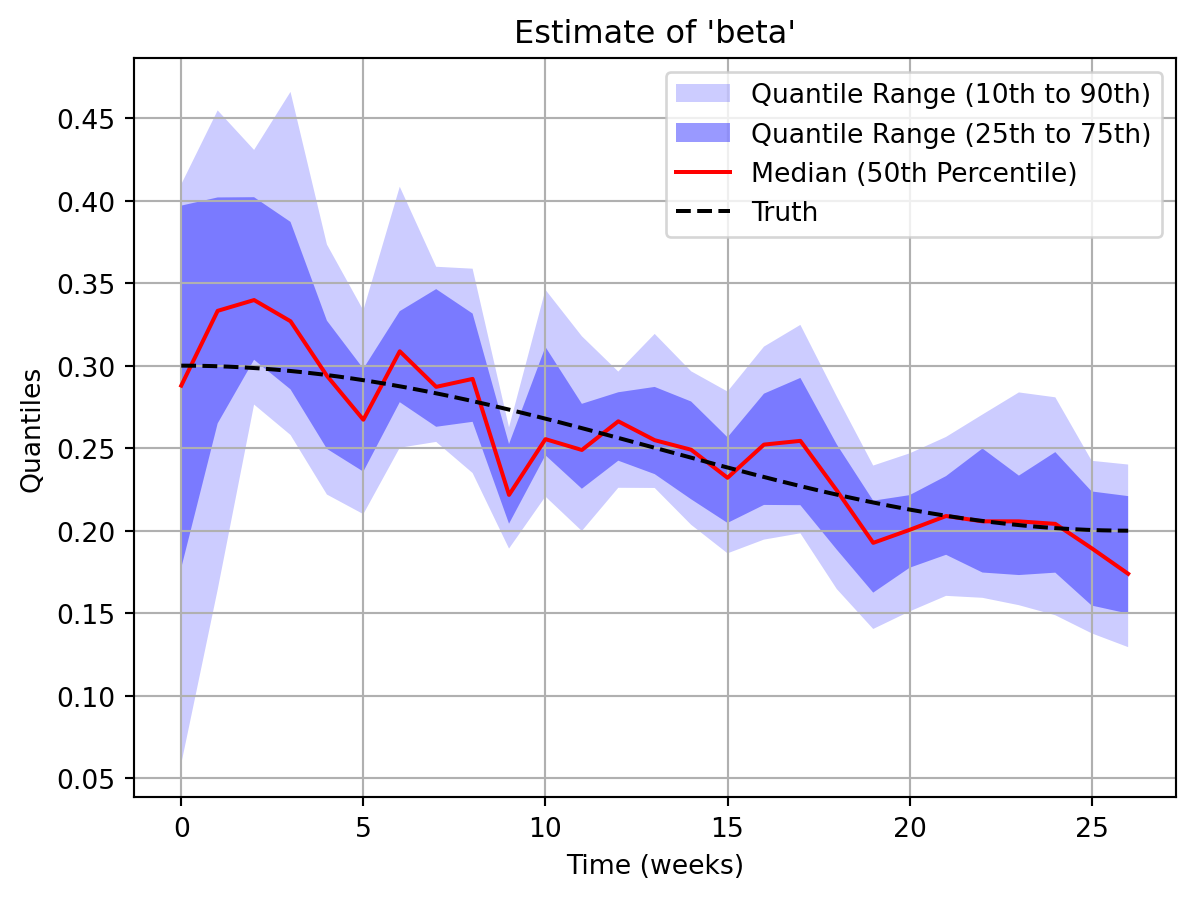
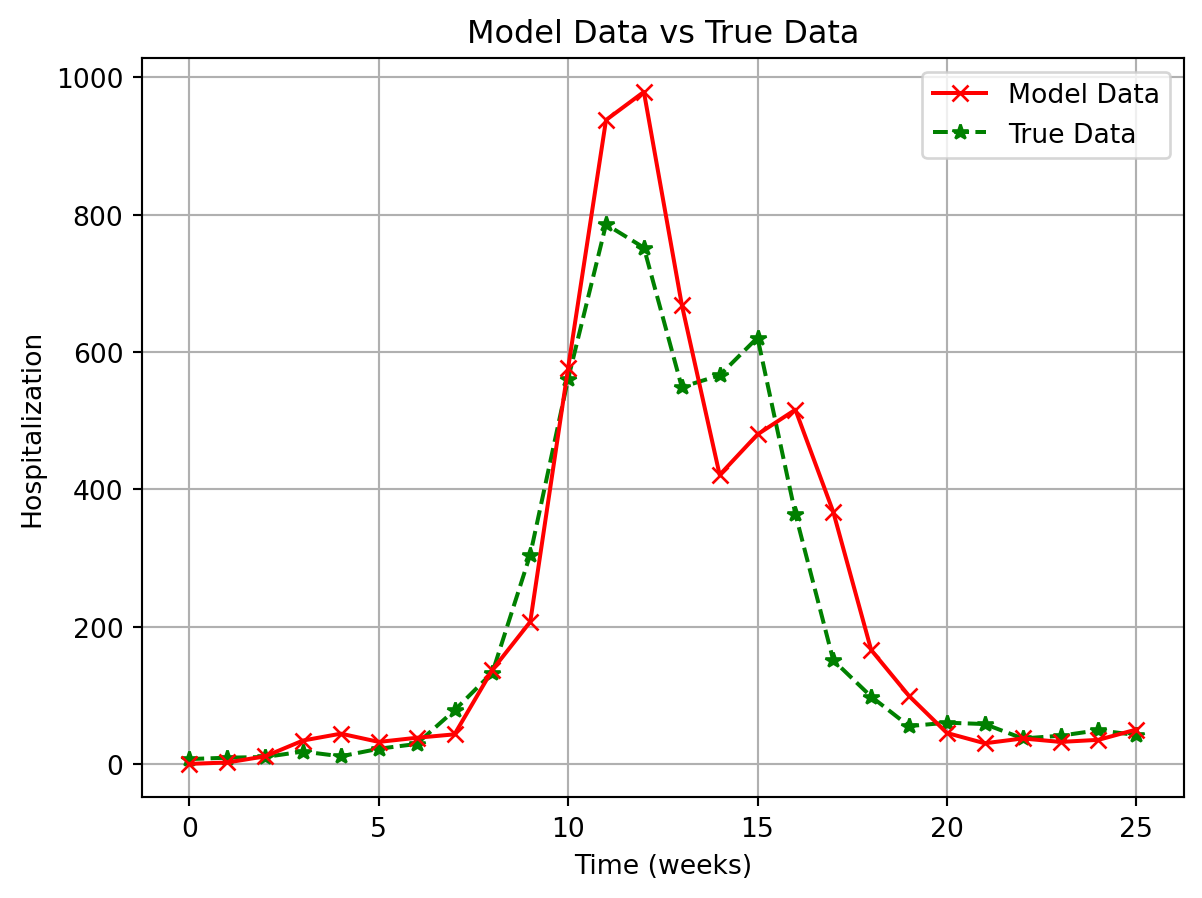
plt.plot(
output.model_data[:, 0],
label="Model Data",
color="red",
linestyle="-",
zorder=10,
marker="x",
)
plt.plot(
output.true_data[:, 0],
label="True Data",
color="green",
linestyle="--",
zorder=9,
marker="*",
)
plt.xlabel("Time (weeks)")
plt.ylabel("Hospitalization")
plt.title("Model Data vs True Data")
plt.legend(loc="upper right")
plt.grid(True)
plt.show()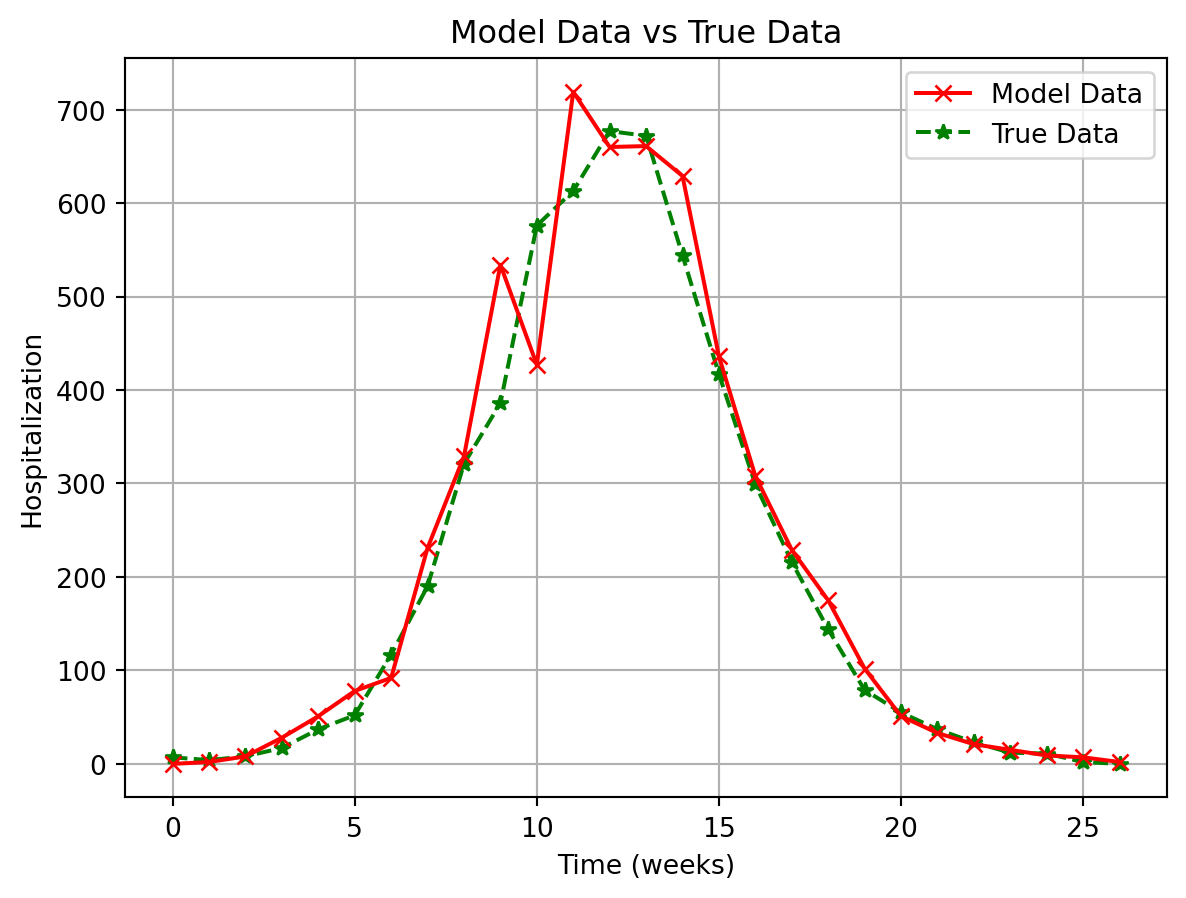
Exercise 3
In this third exercise, we apply the particle filter to real hospitalization data in order to estimate beta. This shows how we can automatically draw data from public sources, in this case the CDC, and link those data to the RUME within epymorph. Because we already tested the setup on synthetic data, we can reuse many of the same objects we defined in the second exercise.
from epymorph.adrio.cdc import InfluenzaStateHospitalization
observations = Observations(
source=InfluenzaStateHospitalization(),
model_link=model_link,
likelihood=Poisson(),
)sim = FilterSimulation(
rume=rume,
observations=observations,
filter_type=filter_type,
params_space=params_space,
)plt.bar(range(len(sim.cases)), sim.cases[:, 0])
plt.title("Arizona Observations from CDC")
plt.xlabel("Time (weeks)")
plt.ylabel("Hospitalizations")
plt.grid(True)
plt.show()
output = sim.run(rng=rng)key = "beta"
node_index = 0
truth = None
key_quantiles = np.array(output.param_quantiles[key])
plt.fill_between(
np.arange(0, len(key_quantiles)),
key_quantiles[:, 3, 0],
key_quantiles[:, 22 - 3, 0],
facecolor="blue",
alpha=0.2,
label="Quantile Range (10th to 90th)",
)
plt.fill_between(
np.arange(0, len(key_quantiles)),
key_quantiles[:, 6, 0],
key_quantiles[:, 22 - 6, 0],
facecolor="blue",
alpha=0.4,
label="Quantile Range (25th to 75th)",
)
plt.plot(
np.arange(0, len(key_quantiles)),
key_quantiles[:, 11, 0],
color="red",
label="Median (50th Percentile)",
)
plt.title(f"Estimate of '{key}'")
plt.xlabel("Time (weeks)")
plt.ylabel("Quantiles")
plt.legend(loc="upper right")
plt.grid(True)
plt.show()
plt.plot(
output.model_data[:, 0],
label="Model Data",
color="red",
linestyle="-",
zorder=10,
marker="x",
)
plt.plot(
output.true_data[:, 0],
label="True Data",
color="green",
linestyle="--",
zorder=9,
marker="*",
)
plt.xlabel("Time (weeks)")
plt.ylabel("Hospitalization")
plt.title("Model Data vs True Data")
plt.legend(loc="upper right")
plt.grid(True)
plt.show()