import matplotlib.pyplot as plt
import numpy as np
from sympy import Max
from epymorph.kit import *
from epymorph.adrio import acs5Birth-death IPM modeling example
SCENARIO:
In many modeling scenarios, it is important to account for changes in host populations over time due to births and deaths. In some cases, losses may be due to fatalities due to pathogen infection; in other scenarios, the organisms being modeled simply have natural life spans or cycles that fall within the modeling timeframe. In either case, we address this issue by adding explicit “birth” and “death” compartments to our model. The exercises in this vignette show you how to do this in epymorph, as well as exploring how birth/death dynamics can affect model behavior.
Exercise 1
Then, add the special compartment called “DEATH” to your model adding several edges from S, I, and R to “DEATH” with a rate of “death_rate*[State Variable]”. It is simplest to assume that birth_rate and death_rate are equivalent.
Note that the model also includes wanining immunity.
class BSIRD_ipm(CompartmentModel):
compartments = [
compartment("S"),
compartment("I"),
compartment("R"),
]
requirements = [
AttributeDef("beta", type=float, shape=Shapes.TxN, comment="infectivity"),
AttributeDef(
"gamma",
type=float,
shape=Shapes.TxN,
comment="progression from infected to recovered",
),
AttributeDef(
"xi",
type=float,
shape=Shapes.TxN,
comment="progression from recovered to susceptible",
),
AttributeDef(
"birth_rate",
type=float,
shape=Shapes.TxN,
comment="birth rate per day per capita",
),
AttributeDef(
"death_rate",
type=float,
shape=Shapes.TxN,
comment="death rate per day per capita",
),
]
def edges(self, symbols):
[S, I, R] = symbols.all_compartments
[β, γ, ξ, br, dr] = symbols.all_requirements
N = Max(1, S + I + R)
return [
# traditional formulation for S,I,R
edge(S, I, rate=β * S * I / N),
edge(I, R, rate=γ * I),
edge(R, S, rate=ξ * R),
# newborn individuals enter S
edge(BIRTH, S, rate=br * (S + I + R)),
# death edges
edge(S, DEATH, rate=dr * S),
edge(I, DEATH, rate=dr * I),
edge(R, DEATH, rate=dr * R),
]Exercise 2
my_ipm = BSIRD_ipm()
my_ipm.diagram()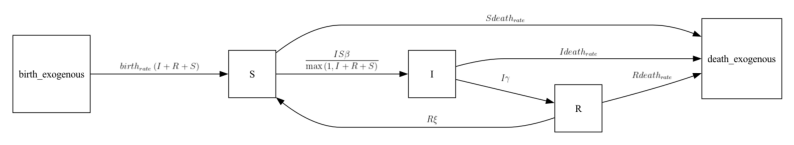
scope = CountyScope.in_counties(["04013"], year=2020) # Maricopa County
time = TimeFrame.of("2020-01-01", 365 * 3)
null_mm = mm.No()my_init = init.SingleLocation(location=0, seed_size=5)rume = SingleStrataRUME.build(
ipm=BSIRD_ipm(),
mm=null_mm,
init=my_init,
scope=scope,
time_frame=time,
params={
"beta": 0.4,
"gamma": 1 / 4,
"xi": 1 / 120,
"birth_rate": 1 / (70 * 365),
"death_rate": 1 / (70 * 365),
"population": acs5.Population(),
},
)
sim = BasicSimulator(rume)
with sim_messaging(live = False):
out = sim.run(
rng_factory=default_rng(5),
)Loading gpm:all::init::population (epymorph.adrio.acs5.Population):
|####################| 100% (0.692s)
Running simulation (BasicSimulator):
• 2020-01-01 to 2022-12-30 (1095 days)
• 1 geo nodes
|####################| 100%
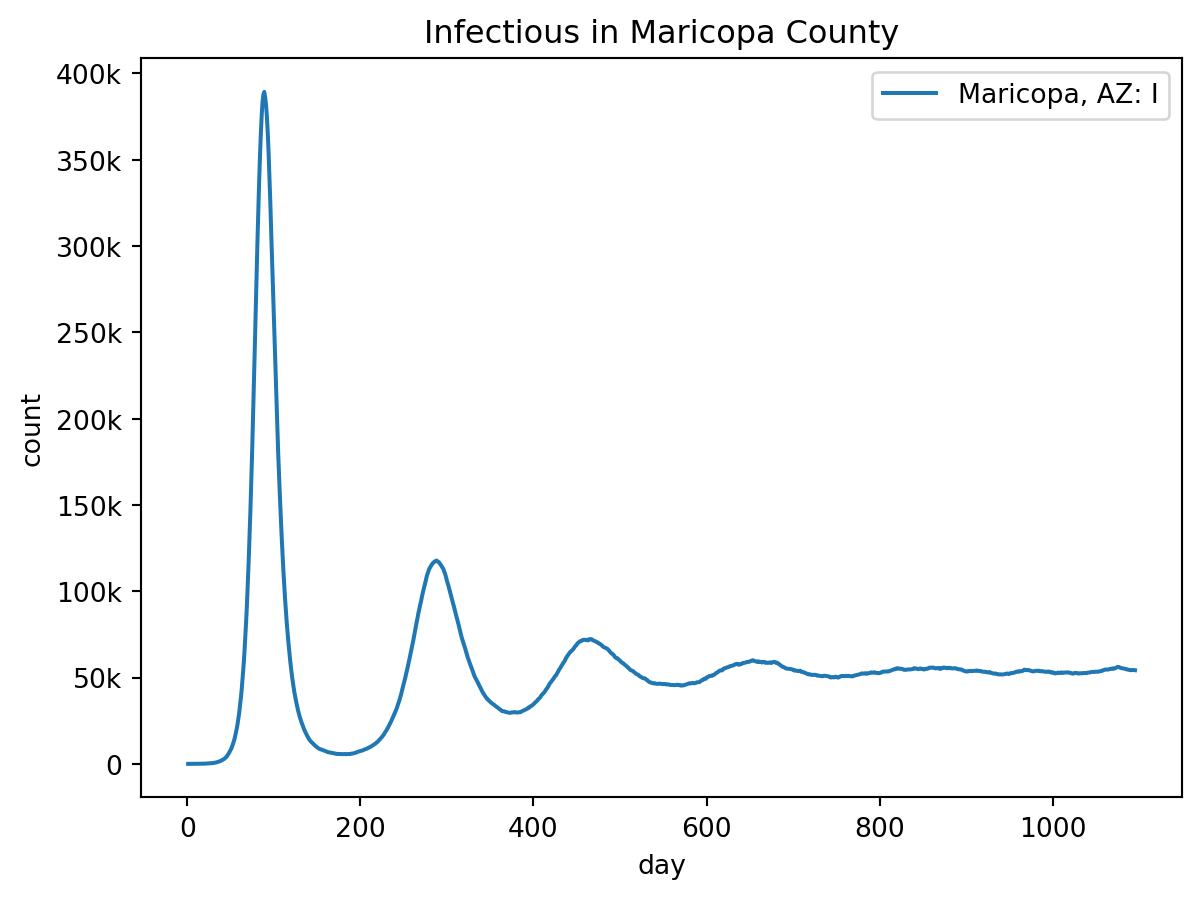
Runtime: 0.296sout.plot.line(
geo=out.rume.scope.select.all(),
time=out.rume.time_frame.select.all(),
quantity=out.rume.ipm.select.compartments("I"),
title="Infectious in Maricopa County",
)
Exercise 3
stored_sim_results = []
initial = [0.01, 0.05, 0.10, 0.15]
for frac in initial:
rume = SingleStrataRUME.build(
ipm=BSIRD_ipm(),
mm=null_mm,
init=init.Proportional(ratios=np.array([1 - frac, frac, 0])),
scope=scope,
time_frame=time,
params={
"beta": 0.4,
"gamma": 1 / 4,
"xi": 1 / 120,
"birth_rate": 1 / (70 * 365),
"death_rate": 1 / (70 * 365),
"population": acs5.Population(),
},
)
sim = BasicSimulator(rume)
out = sim.run(
rng_factory=default_rng(5),
)
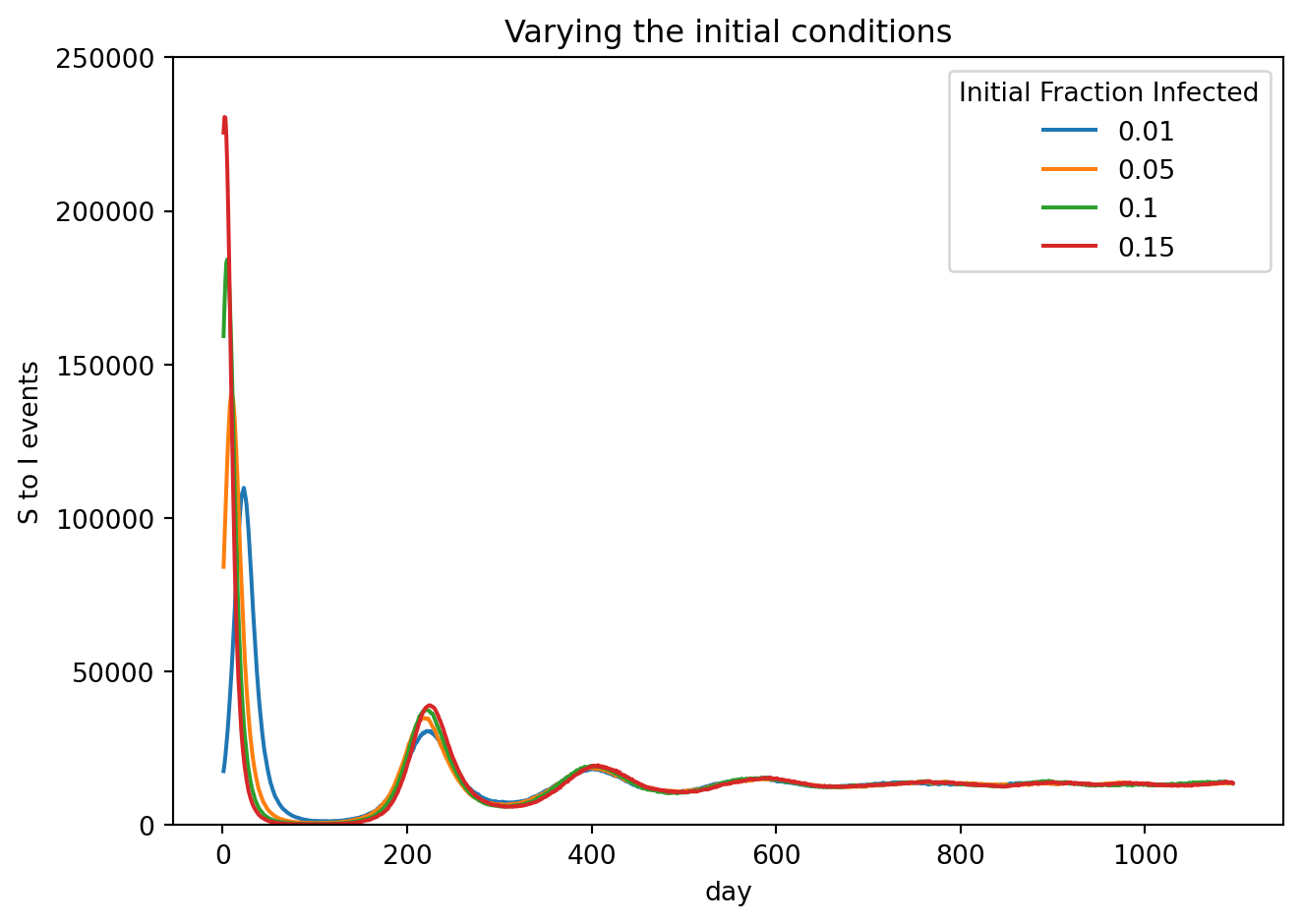
stored_sim_results.append(out)fig, ax = plt.subplots()
for j in stored_sim_results:
j.plot.line_plt(
ax = ax,
geo=j.rume.scope.select.all(),
time=j.rume.time_frame.select.all(),
quantity=j.rume.ipm.select.events("S->I"),
)
ax.set_ylim(bottom=0,top=2.5e5)
ax.set_xlabel("day")
ax.set_ylabel("S to I events")
ax.set_title("Varying the initial conditions")
ax.grid(False)
plt.legend(initial, title="Initial Fraction Infected")
fig.tight_layout()
plt.show()